Background: CLL is the most prevalent adult leukemia with a highly heterogenous genetic landscape leading to extremely varied clinical outcomes. Previously, CLL patients with immunoglobulin heavy chain variable region ( IGHV) mutated status and presence of deletion 13q14 [del(13q)] were identified as favorable-risk with most patients remaining on lifetime observation. However, other intrinsic factors contributing to CLL disease progression and leading to treatment initiation remain unknown. Our goal was to identify patients at high-risk of progression with added predictive value (APV) beyond IGHV-status and presence of del(13q).
Methods: Flow cytometry was used to sort CD19+CD5+ malignant cells in peripheral blood from CLL patients at Huntsman Cancer Institute at University of Utah and triple sequencing generated (tumor whole transcriptome and paired tumor-normal whole exome data). We used SPECTRA, a novel dimension reduction transcriptome approach, to derive a set of independent, intrinsic, quantitative transcriptome tumor variables from 249 CLL patients with quality-controlled transcriptome sequence data. From this set, 182 treatment-naïve (TN) patients with quality-controlled clinical variables were used as a discovery cohort set to derive a SPECTRA-based risk score for TTFT using multivariable Cox proportional hazards modeling. Event time was date of patient specimen acquisition to initiation of frontline therapy with patients censored at last follow-up or death. Gaussian mixture modeling (GMM) was performed to identify evidence for distinct risk groups within the quantitative risk score. GMM-risk groups, and further stratifications by IGHV mutational and del(13q) status, were used in hazard ratios (HR) and to illustrate differences in Kaplan-Meier time-to-event curves. An independent publicly available dataset of 548 TN CLL patients with clinical and transcriptome data for CD19+ enriched cells was used for replication.
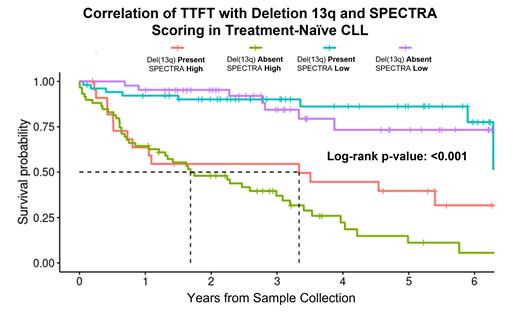
Results: SPECTRA produced 19 quantitative spectra variables to describe most of the expression variation across the 249 CLL patients. In multivariable Cox modeling for TTFT, 7 spectra were significant and retained in a final model, which GMM separated into a bi-modal distribution and defined high (N=83) and low-risk (N=99) groups (HR=5.7, 95% confidence interval [CI] 3.4-9.7). In a post-hoc model including IGHV mutational and del(13q) status, the spectra-risk score showed significant APV beyond both factors (p<0.001 for both). The discovery study timeframe was 6.3 years (y) and included 109 IGHV-mutated and 75 del(13q) patients. In the IGHV-mutated patients, median TTFT (mTTFT) was significantly shorter for spectra-high risk patients (3.1y) vs. spectra-low risk patients (median not reached [NR]; p<0.001). Similarly, in the del(13q) patients, mTTFT was significantly shorter in high-risk patients (3.3y) compared to median NR for spectra-low (p<0.001) ( Figure). Importantly, stratification of patients into low- and high-risk SPECTRA scores provided significant additional information for prediction of TTFT than IGHV mutational and del(13q) status ( Figure). For validation, we applied the same spectra-risk score to the independent data set containing 301 IGHV-mutated and 278 del(13q) CLL patients. In the replication set, the mTTFT in the IGHV-mutated patients who were spectra-high was 3.3y vs. 11.2y in spectra-low (p<0.001). Similarly, in the replication set of del(13q) patients, spectra-high-risk patients had a mTTFT of 2.7y vs. 10.7y for spectra-low (p<0.001).
Conclusion: A transcriptome risk score utilizing novel SPECTRA variables identified a substantial subset of CLL patients with IGHV-mutated status and presence of del(13q) who have uncharacteristically short TTFT, but who would be classified as favorable risk under current best practice stratification. Future directions include validating our results with external patient cohorts from Mayo Clinic and investigating the genetic and transcriptomic profile of SPECTRA low- and high-risk cohorts to determine targetable pathways for treatment.
Disclosures
Feusier:ARUP Laboratories: Current Employment. Stephens:AbbVie: Consultancy; AstraZeneca: Consultancy, Research Funding; BeiGene: Consultancy; Bristol-Myers Squibb: Consultancy; Celgene: Consultancy; Genentech: Consultancy; Janssen: Consultancy; Lilly: Consultancy; Novartis: Research Funding.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal